Search from website
Search from website
Home COVID-19
BioBlock® nasal spray is a natural product derived from bovine colostrum that is meant to serve as a prophylactic measure, creating a protective environment on the nasal mucosa, thereby isolating the viral particles and help prevent the spread of the SARS-CoV-2 virus. It’s meant to be an additional preventive measure wherever it is difficult to keep distance from each other. It has been available for sale in Estonia market since May 2021.
Icosagen has analyzed the ability of the antibodies, isolated from immunized bovine colostrum, to inhibit the binding of the viral spike protein and human cellular receptor ACE2. Biochemical analysis and a pseudovirus assay showed that colostrum derived immunoglobulins from immunized cows neutralize pseudovirus displaying different Spike proteins of currently circulating SArS-CoV-2 variants.
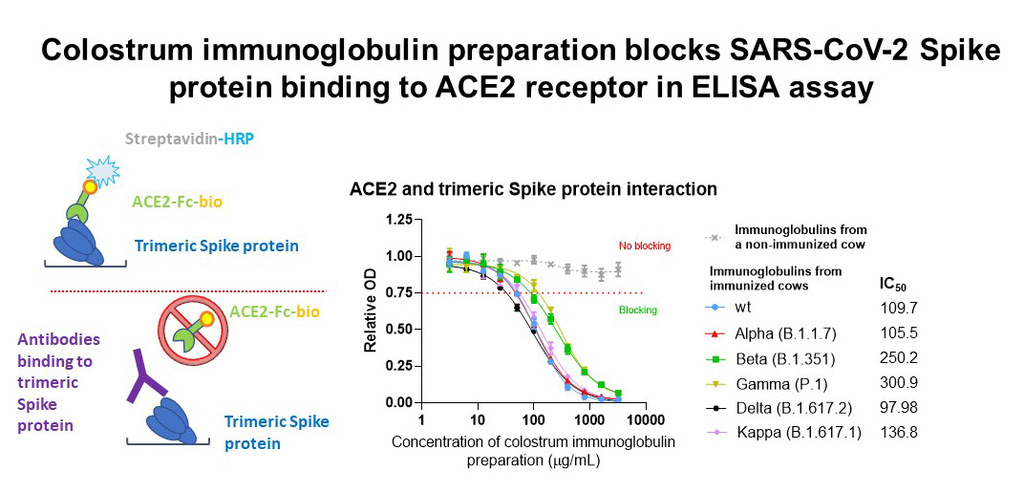
In biochemical analysis, the SARS-CoV-2 trimeric spike protein was attached to the assay plates and the antibodies used in BioBlock® were added. The antiviral antibodies in the solution will bind to the spike protein at the bottom of the plate. Then, ACE2 receptors were added, which were unable to bind to the viral spike protein when the previously added antibodies block it. The amount of binding between the ACE2 receptor and the viral spike protein is measurable, and the lower the binding, the better the added antibodies block the entry of the virus.
To further characterize the virus neutralizing potency of the antibodies, the pseudovirus neutralization assay was performed.
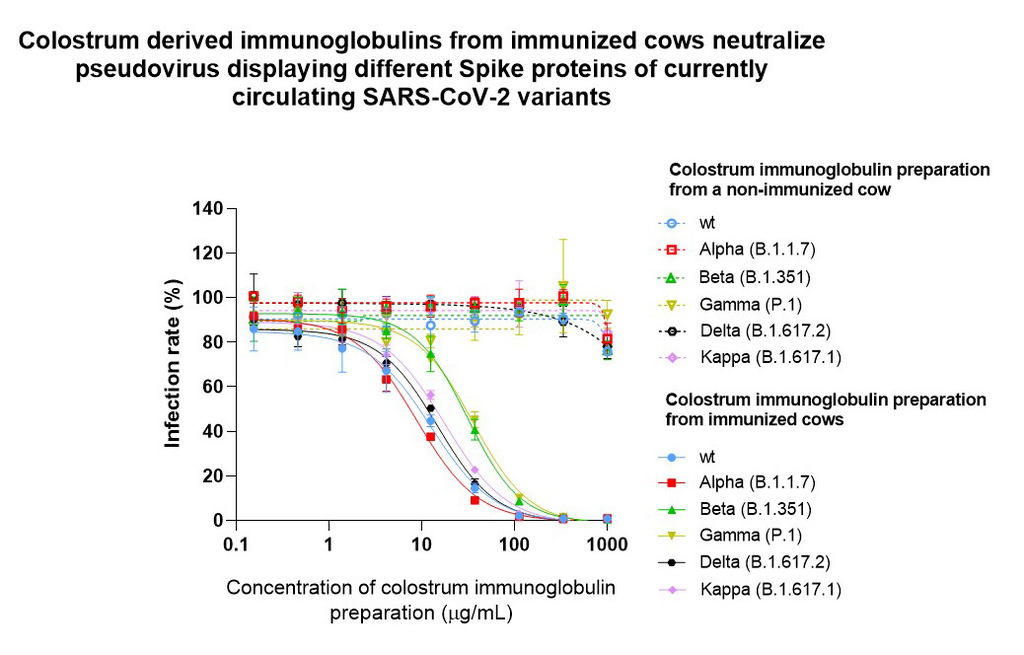
In a pseudovirus neutralization assay, the ability of the viral particle to bind ACE2 receptor and entry to cells, is directly measured. Compared to biochemical analysis, this assay is even more similar to an actual viral infection as it allows to measure the efficacy of spike protein-dependent infection and inhibition of viral entry into the cell occurs as decreased enzymatic activity. It is important to note that this effect was entirely based on specific antiviral antibodies produced by immunization, as colostrum antibodies from non-immunized cows showed no effect.
Our results so far show that colostrum derived immunoglobulins from immunized cows block the interaction between the Spike protein and ACE2 receptor from five different virus strains in a concentration-dependent manner, thus blocking the virus from entering the host cells.
BioBlock was an achievement of consortium, which includes Icosagen Cell Factory OÜ, AS Chemi-Pharm, OÜ Teadus ja Tegu, Estonian University of Life Sciences (Institute of Veterinary Medicine and OÜ Eerika farm) and University of Tartu (Institute of Pharmacy, Institute of Biomedical and Translational Medicine, and Institute of Technology).
For companies, that wish to purchase BioBlock® nasal spray for their employees, please contact: sales@icosagen.com, phone: +372 5857 7040.
The U.S. Patent Office registered and filed OÜ Icosagen Cell Factory’s Patent Application No. 63090576 „SARS-CoV-2 Neutralizing Antibodies“ on October 12, 2020. Thus, Icosagen has successfully completed the first phase of the development of biological drugs against Covid-19, virus-neutralizing antibodies. With this patent application, Icosagen Cell Factory OÜ wishes to obtain intellectual property protection for new antiviral therapeutic antibodies in the USA. Next, the company plans to implement the intellectual property protection around the world.
The patent discloses four antibodies against SARS-CoV-2 based on the sequences of B-lymphocyte genes isolated from the blood of Covid-19 recovered donors. Cells with anti-SARS-CoV-2 antibodies were screened and the genetic sequence encoding antibodies were isolated using Icosagen’s previously patented HybriFree technology. To increase the efficiency of the resulting antibodies, the amino acid sequences were optimized and used to generate IgG antibody isotype. Most potent variations were produced using Icosagen’s QMCF Technology. Based on the biochemical and biophysical parameters of the antibodies, it can be stated that Icosagen has been able to isolate high-affinity antibodies against the SARS-CoV-2 viral spike protein (S protein).
On September 3rd the Health Board registered our developed diagnostic test with the IVD CE mark. The test can be used to detect antibodies against SARS-CoV-2 in human blood samples to determine if a person has had the disease. Volunteers recovered from COVID-19, whose donated blood was used to validate the test, played a very important role in the validation of the test. Long-term studies are underway to determine the duration of antiviral immunity associated with COVID-19 and possible protection against future infections. The test is based on recombinant viral proteins produced by Icosagen and are also available in our online catalogue. The test works by enzyme-linked immunosorbent assay (ELISA), allowing the semi-quantitative in vitro detection of IgG and IgM antibodies against SARS-CoV-2 virus in human serum or plasma (Medical Device Database (MSA) code 15531).
"The validation of the COVID-19 ELISA test is an important milestone in our company’s development, which we have reached through the joint efforts of Icosagen staff, volunteer donors and partners, such as the Government of Estonia, which contacted us to provide assistance." commented Mart Ustav, founder and CEO of Icosagen. "We are no longer only a subcontractor to produce bolts and nuts for medical devices, but we are now able to develop, clinically validate and produce diagnostic tests. We do not intend to dwell here satisfactorily, but are also developing the SARS-CoV-2 antigen test and the COVID-19 neutralizing antibody ELISA test."
The SARS-CoV-2 antigen assay in development allows the detection of SARS-CoV-2 virus in a patient's saliva sample at an early stage of infection, providing a rapid and cost-effective alternative to the current RNA assay. And the second test in development — the COVID-19 neutralizing antibody ELISA — can specifically detect the presence of virus neutralizing antibodies in patients' blood serum and makes it possible to determine the strength and duration of the immunity elicited by the vaccine. In addition, this test allows medical authorities to select highly neutralizing sera from the blood sera of patients who have recovered from the disease, for use in the treatment of critically ill patients.
The Ministry of Social Affairs of the Republic of Estonia supported the creation of the newly approved tests, which are still under development, with 100,000 euros.
In response to COVID-19 pandemia, we have produced and purified recombinant SARS-CoV-2 antigens from CHO cells for diagnostic use in gram quantities. Multiple grams of N, S1, S2 and RBD proteins will be produced with stable CHO cell lines that are currently under development. These viral proteins are ideal for use as antigens in antibody discovery or as immunological tools to measure the concentration of virus-specific IgG, IgM or IgA antibodies in patient’s blood samples.
Table 1: Antigens currently available for purchase
Nucleoprotein
Spike S1
RBD
RBD2
Spike S2
ACE2
ACE2-Fc
Production cell line
CHO
CHO
CHO
CHO
CHO
CHO
CHO
Molecular Weight (kDa)
46.9
76.4
23
26.4
58.9
84.9
110
Purity
>90%
> 90%
>95%
>95%
> 90%
>80%
>95%
Amino acids
Full length
14-681
331-524
319-541
693-1218
20-742
18-740
Glycosylated
Yes
Yes
Yes
Yes
Yes
Yes
Yes
Binding to ACE2
NA
Yes
Yes
Yes
Yes
NA
NA
Tested on clinical samples
Yes
Yes
Yes
Yes
Yes
NA
NA
SARS-CoV-2 is a positive-sense single-stranded RNA virus that can cause severe respiratory illness COVID-19. It is a strain of severe acute respiratory syndrome-related coronavirus, and its 30 kb genome encodes four structural proteins — S (Spike), E (Envelope), M (Membrane) and N (nucleocapsid).
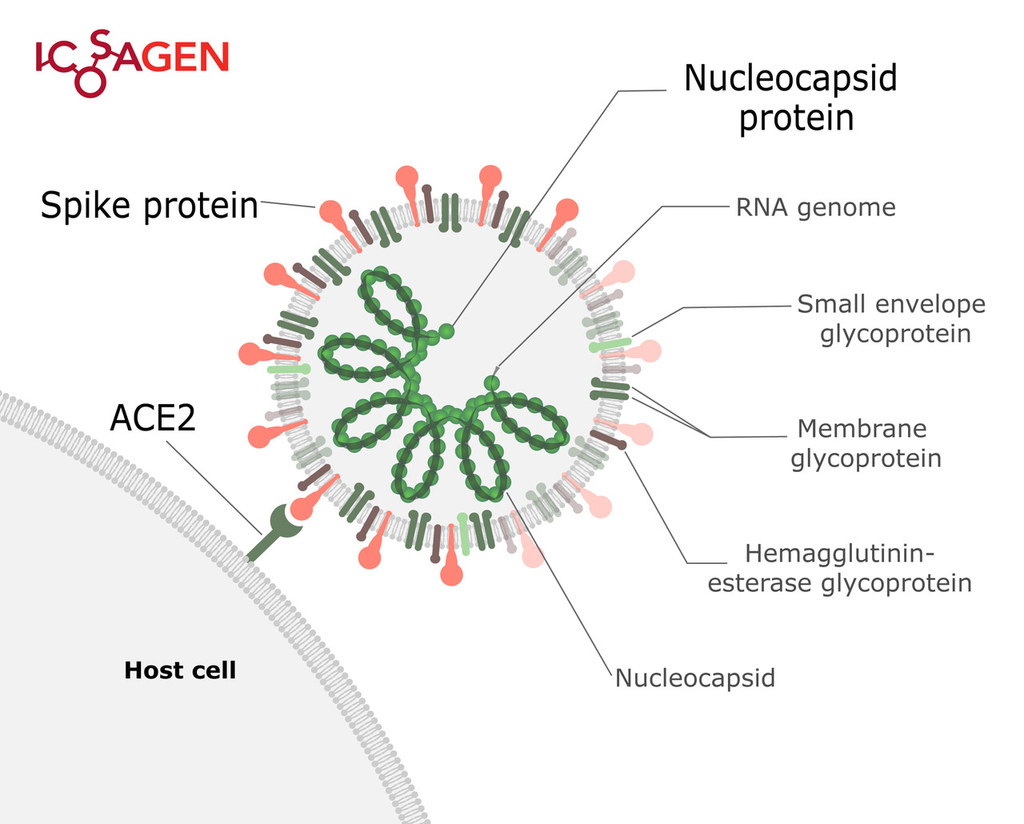
Figure 1. Structure of SARS-CoV-2.
Spike (S) protein is a trimeric viral surface protein consisting of two subunits, S1 and S2. The RBD (receptor binding domain) of S1 subunit binds to the ACE2 receptor. Neutralizing antibody response to the S protein plays an important role in immunity. Homotrimers of the S protein make up the distinctive spike-like structure that is heavily N-glycosylated.
The nucleocapsid protein (N) is responsible for packaging and protecting coronavirus genomic RNA. It is a good target for virus detection as nucleocapsid protein is the most abundant protein of coronavirus and it is highly immunogenic.
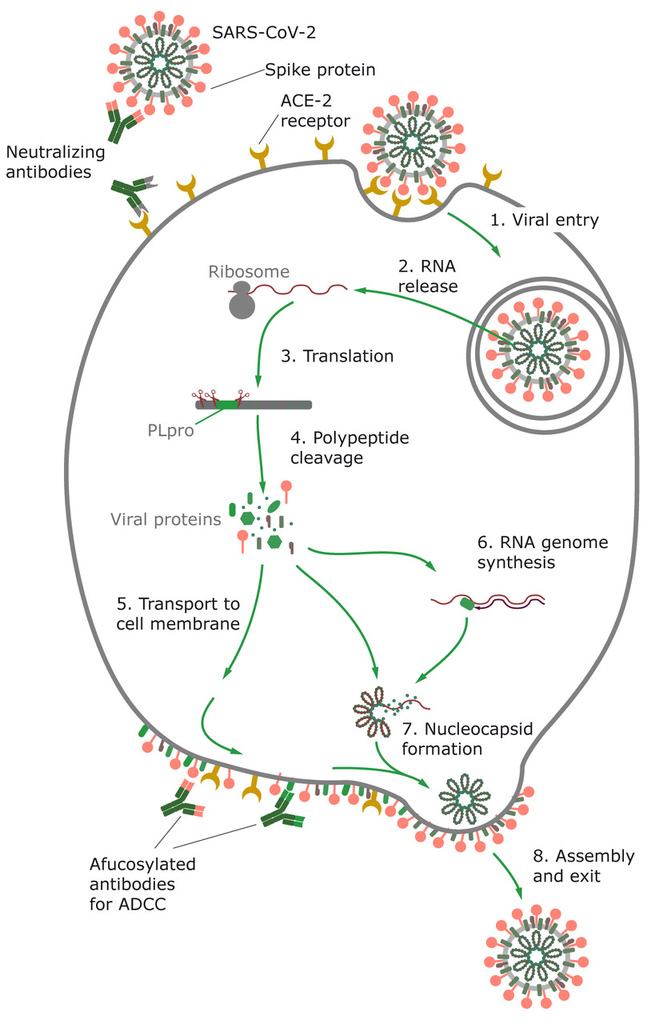
Figure 2. SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) receptor to enter the host cell, where the virus replicates. Virus inserts its RNA into the host cell, where it forces the cell to produce new virus proteins and replicates the RNA genome. Viral proteins are glycosylated and transported to the host cell membrane, where the assembled particles bud from the host cell.
Using our patented QMCF technology and vast experience in protein production, we have produced and purified functional recombinant SARS-CoV-2 antigens from CHO cells for diagnostic use. All proteins go through rigorous testing to ensure the highest quality and similarity to natural protein forms. For Spike S1, RBD and RBD2 binding to ACE-2 has been confirmed using the Octet platform.
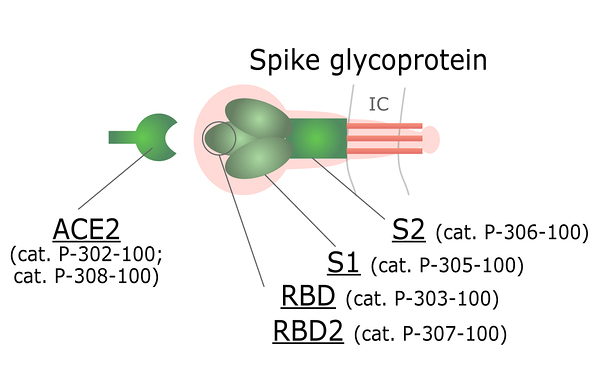
Figure 3. ACE2 and different domains of Spike protein available for purchase.
All the antigens are produced in CHO cells, which enables proper glycosylation pattern and thus contributes to protein’s antigenic properties. PNGase F treatment displays the extent of N-glycosylation, where a large mobility shift of N and S proteins is seen when comparing deglycosylated and native forms, indicating that these proteins are N-glycosylated.
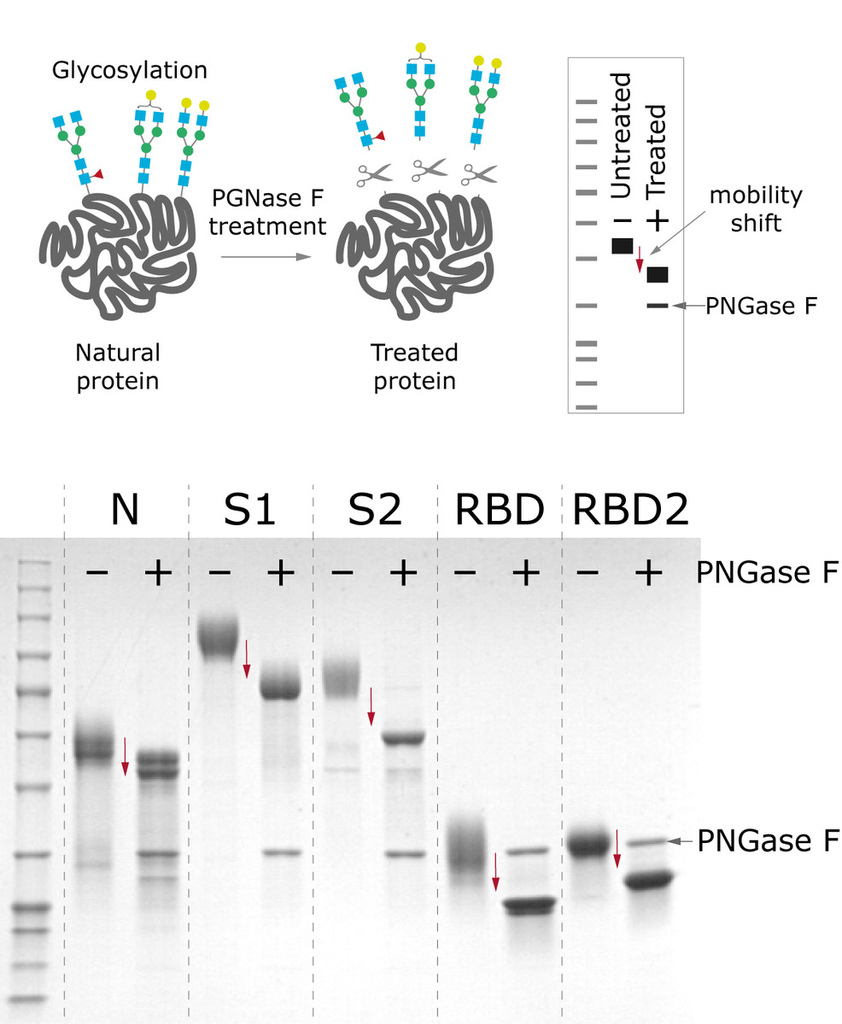
Figure 4. Mobility shift between produced antigens before and after deglycosylation treatment with PNGase F.
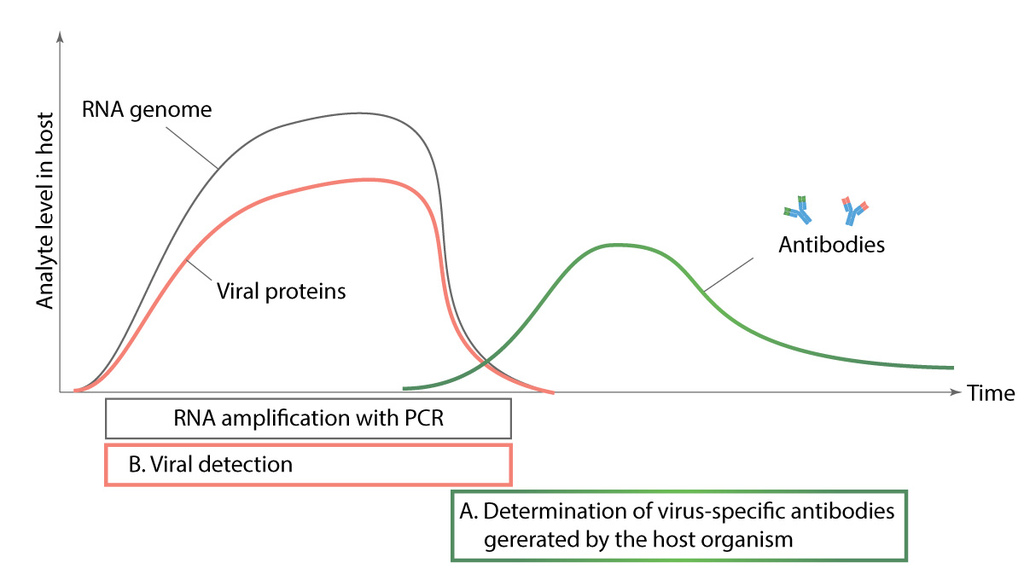
Figure 5. Comparison of different diagnostic methods.
Diagnostic tests for detecting SARS-CoV-2 infection can be divided into three- nucleic acid analysis, serological antibody identification and virus detection with diagnostic antibodies. We are focused on developing the two latter named diagnostic tools. Nucleic acid analysis is based on identifying RNA of the virus during infection from nasopharyngeal swab samples, while serological tests look for antibodies generated against virus proteins (antigens). Diagnostic antibodies are used to detect viral proteins from nasopharyngeal swab samples. Nucleic acid analysis and diagnostic antibodies are used to detect active infection while serological tests show us if patient has already had the virus.Figure 5. Comparison of different diagnostic methods (JOONIS)
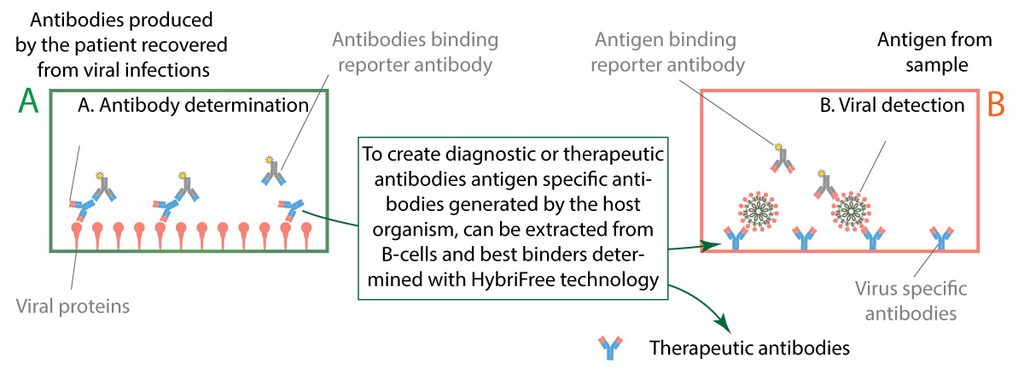
Figure 6. Two types of immunoassay diagnostic tests.
We are closely collaborating with Estonian health system to gather clinical samples from patients recovered from COVID-19. Our serological tests are functional and we are in the process of validating with mentioned clinical samples. Clinical samples also provide vital information about the characteristics of immune response against SARS-CoV-2 infection, showing which antigens are targeted more often and which of IgG, IgA and IgM are more common. From patients’ blood samples we have extracted antibodies binding to viral antigens and are currently screening for most effective diagnostic and therapeutic antibodies.
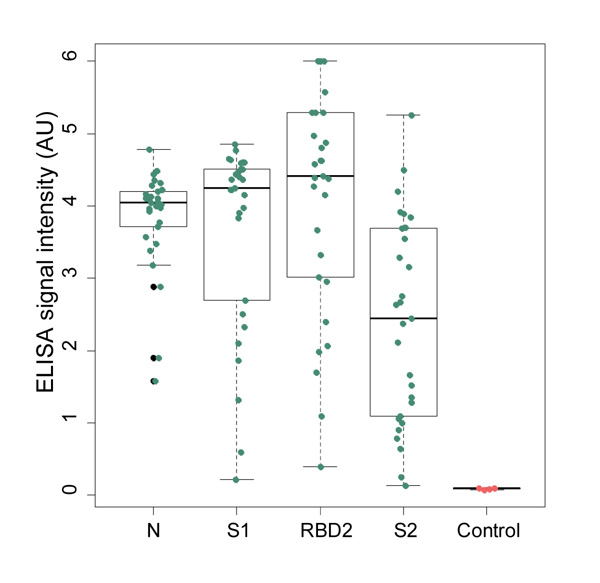
Figure 7. N, S1, S2 and RBD proteins’ ability to capture specific IgG antibodies from clinical samples has been experimentally confirmed with ELISA assay.



