Search from website
Search from website
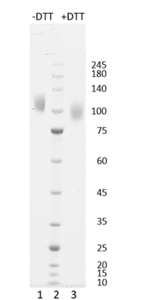
Figure 1. Coomassie-stained SDS-PAGE analysis of SARS-CoV-2 Spike S1 C.37. 4-12% gradient gel is used for analysis. Lane 1. 0.8 µg SARS-CoV-2 Spike S1 C.37 (-DTT). Lane 2. Protein marker (Smobio). Lane 3. 0.8 µg SARS-CoV-2 Spike S1 C.37 (+DTT).
Figure 2. HPLC analytical SEC for final product.
Figure 3. HPLC analytical SEC after 3 freeze-thaw cycles.
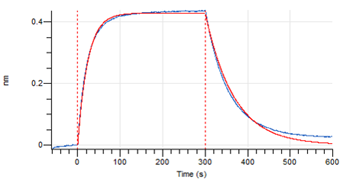
Figure 4. Octet RED96e analysis of SARS-CoV-2 Spike S1 C.37 binding to the ACE2 receptor.
Catalogue #
P-358-100
Name
SARS-CoV-2 Spike S1 C.37 (lambda)
Description
Protein contains amino acids 14-681, mutations G75V, T76I, del 246-252, L452Q, F490S, D614G, two extra amino acids (AS) in N-terminus and His-6 tag at C-terminus and GSG linker between protein and tag.
Uniprot ID
P0DTC2
MW
75.1 kDa
Host
CHO-based cell line (expressed by QMCF Technology)
Purification
Purified by Ni-affinity chromatography and gel-filtration from serum-free CHO growth media, sterile filtrated
Concentration
1 mg/ml
Buffer
PBS pH 7.4
Endotoxine
NA
QC
SDS-PAGE, NanoDrop A280, Analytical SEC, Octet binding to ACE2
Shipping
Shipped on dry ice.
Storage
Store at -70°C upon receipt. Recommended to aliquot into smaller quantities. Avoid repeated freeze-thaw cycles
This product is for research use only
TECHNICAL ASSISTANCE
Please refer any technical questions to
technical.support@icosagen.com